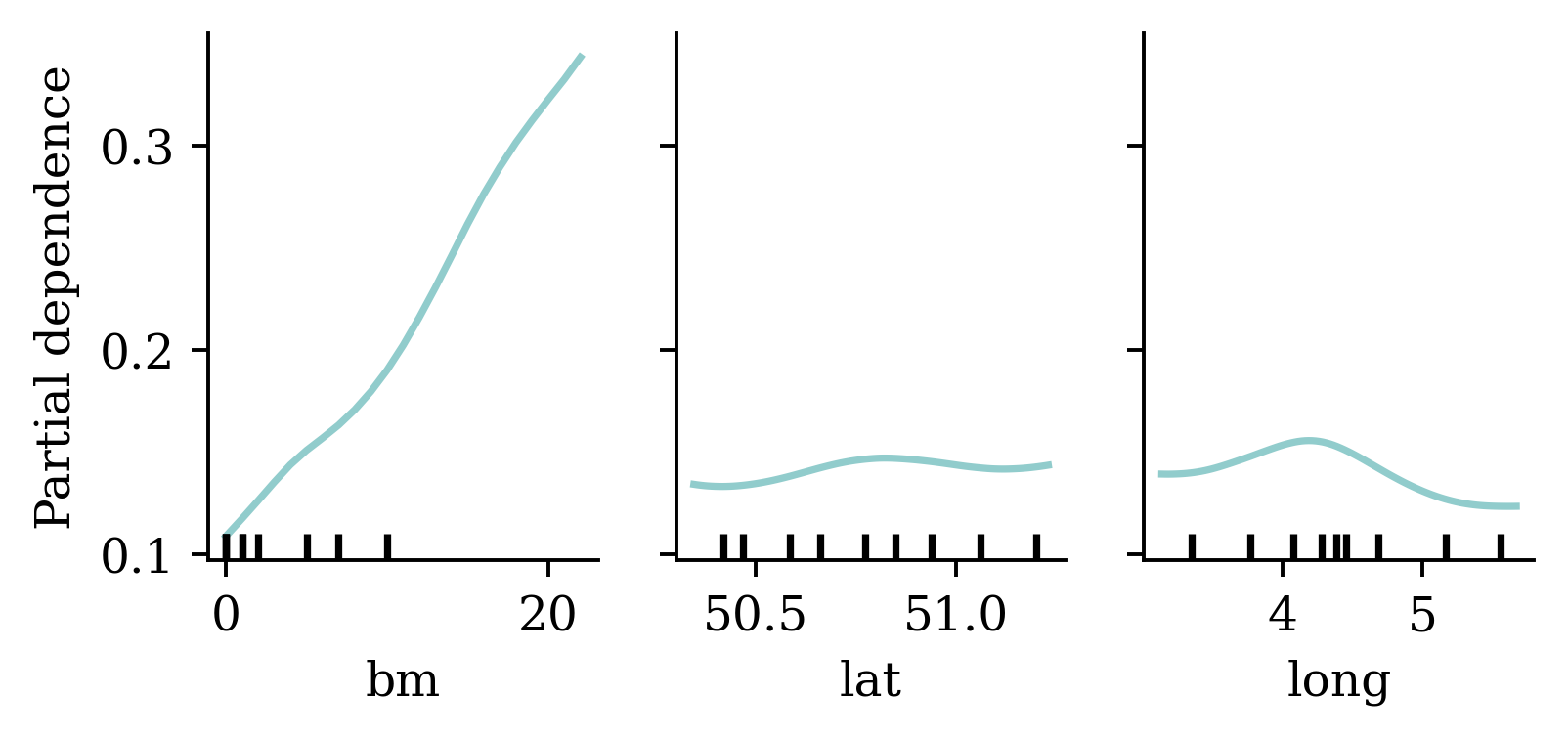
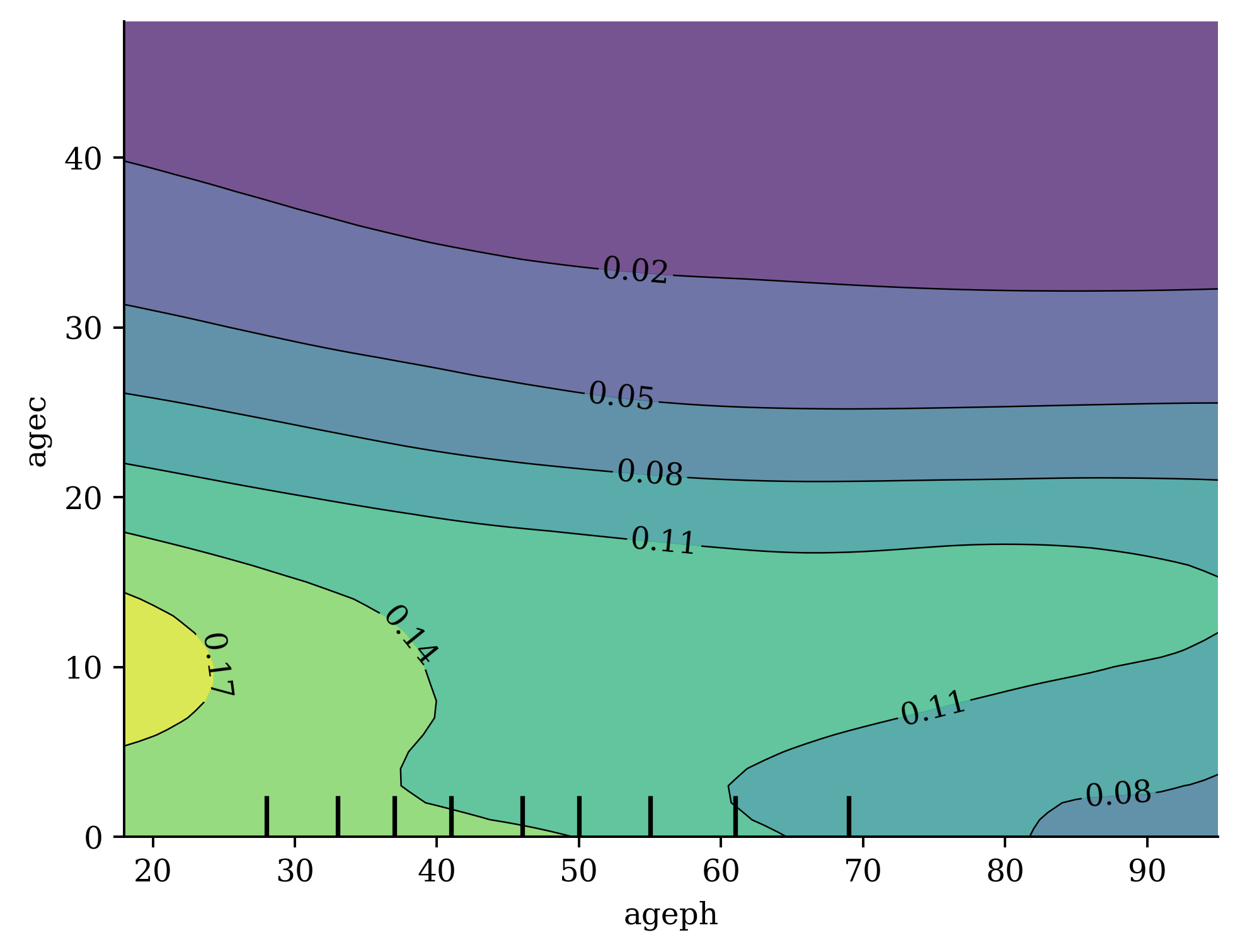
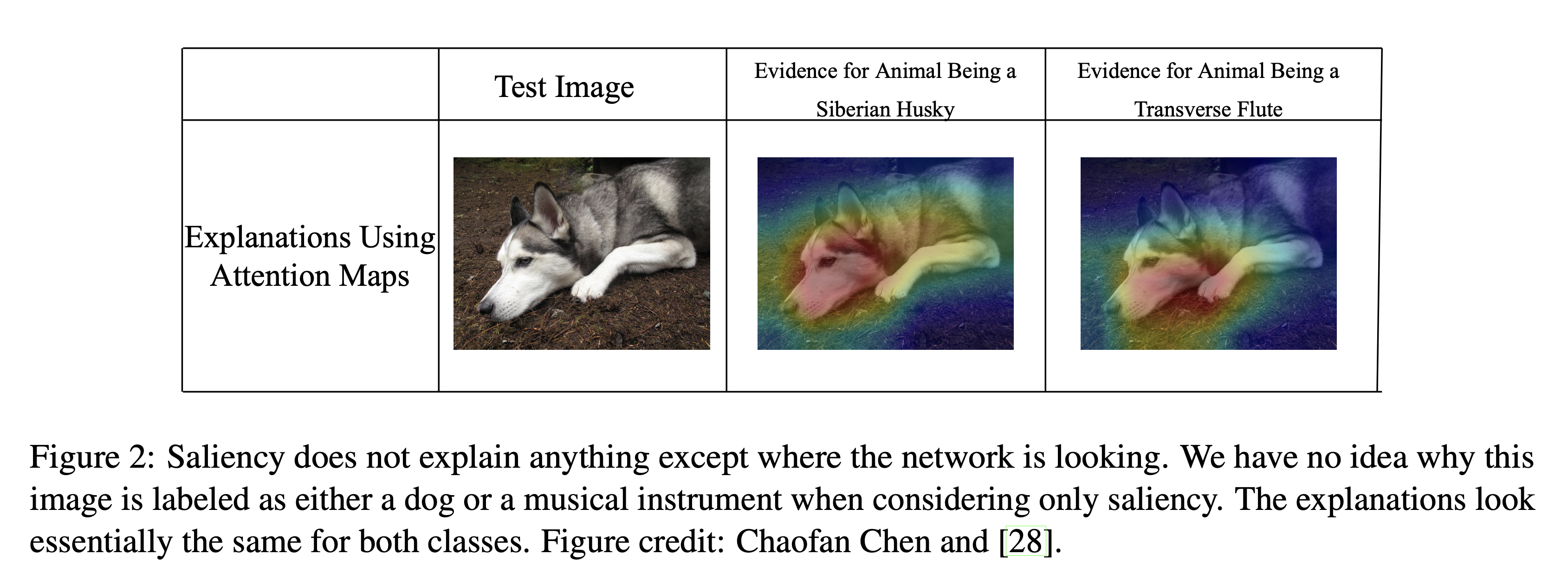
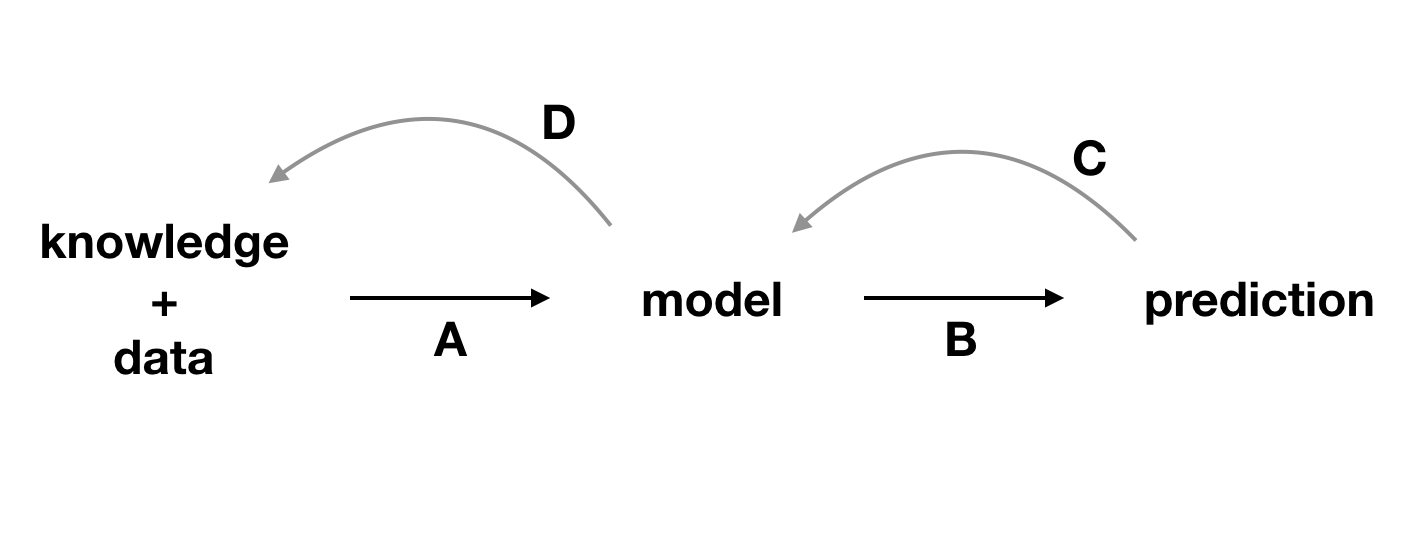
Ben-Zion, Ziv, Kristin Witte, Akshay K Jagadish, Or Duek, Ilan Harpaz-Rotem, Marie-Christine Khorsandian, Achim Burrer, et al. 2025. “Assessing and Alleviating State Anxiety in Large Language Models.” Npj Digital Medicine 8 (1): 132.
Biecek, Przemyslaw, and Tomasz Burzykowski. 2021.
Explanatory Model Analysis. Chapman; Hall/CRC, New York.
https://pbiecek.github.io/ema/.
Breiman, Leo. 2001. “Random Forests.” Machine Learning 45 (1): 5–32.
Carreira-Perpinán, Miguel A, and Pooya Tavallali. 2018. “Alternating Optimization of Decision Trees, with Application to Learning Sparse Oblique Trees.” Advances in Neural Information Processing Systems 31.
Charpentier, Arthur. 2024. Insurance, Biases, Discrimination and Fairness. Springer.
Chen, Zhanhui, Yang Lu, Jinggong Zhang, and Wenjun Zhu. 2024. “Managing Weather Risk with a Neural Network-Based Index Insurance.” Management Science 70 (7): 4306–27.
Delcaillau, Dimitri, Antoine Ly, Alize Papp, and Franck Vermet. 2022. “Model Transparency and Interpretability: Survey and Application to the Insurance Industry.” European Actuarial Journal 12 (2): 443–84.
Doshi-Velez, Finale, and Been Kim. 2017. “Towards a Rigorous Science of Interpretable Machine Learning.”
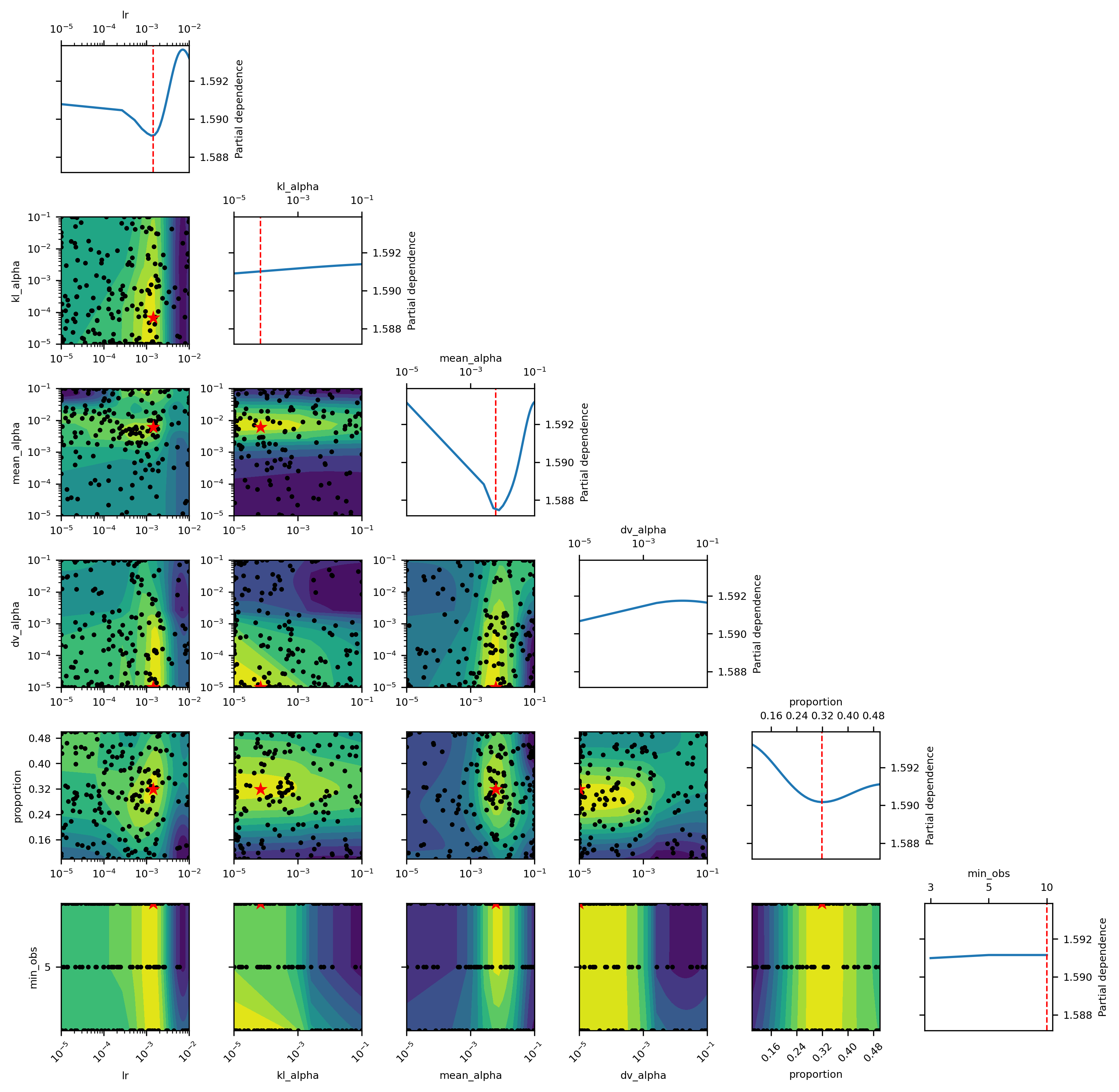
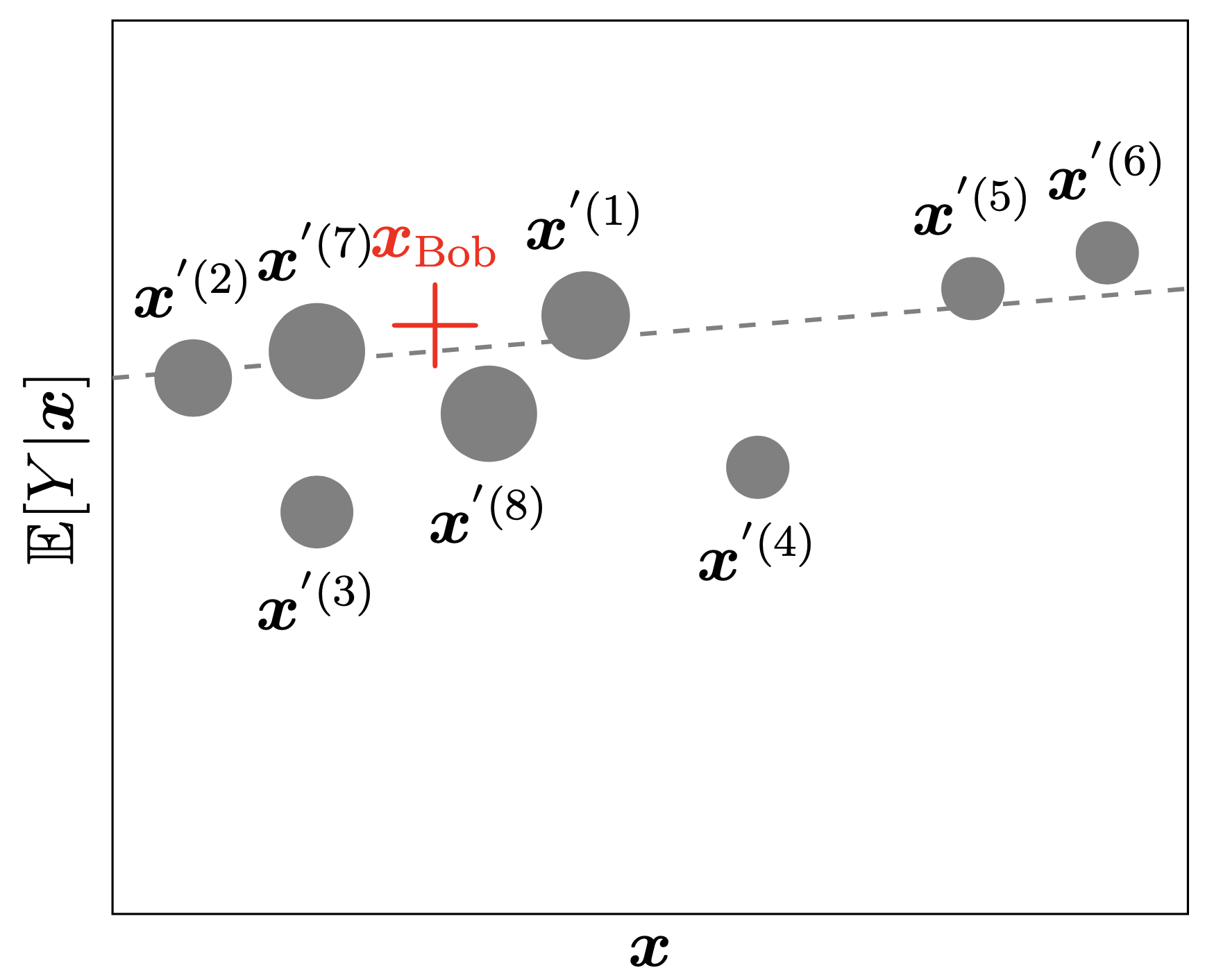
Fisher, Aaron, Cynthia Rudin, and Francesca Dominici. 2019. “All Models Are Wrong, but Many Are Useful: Learning a Variable’s Importance by Studying an Entire Class of Prediction Models Simultaneously.” Journal of Machine Learning Research 20 (177): 1–81.
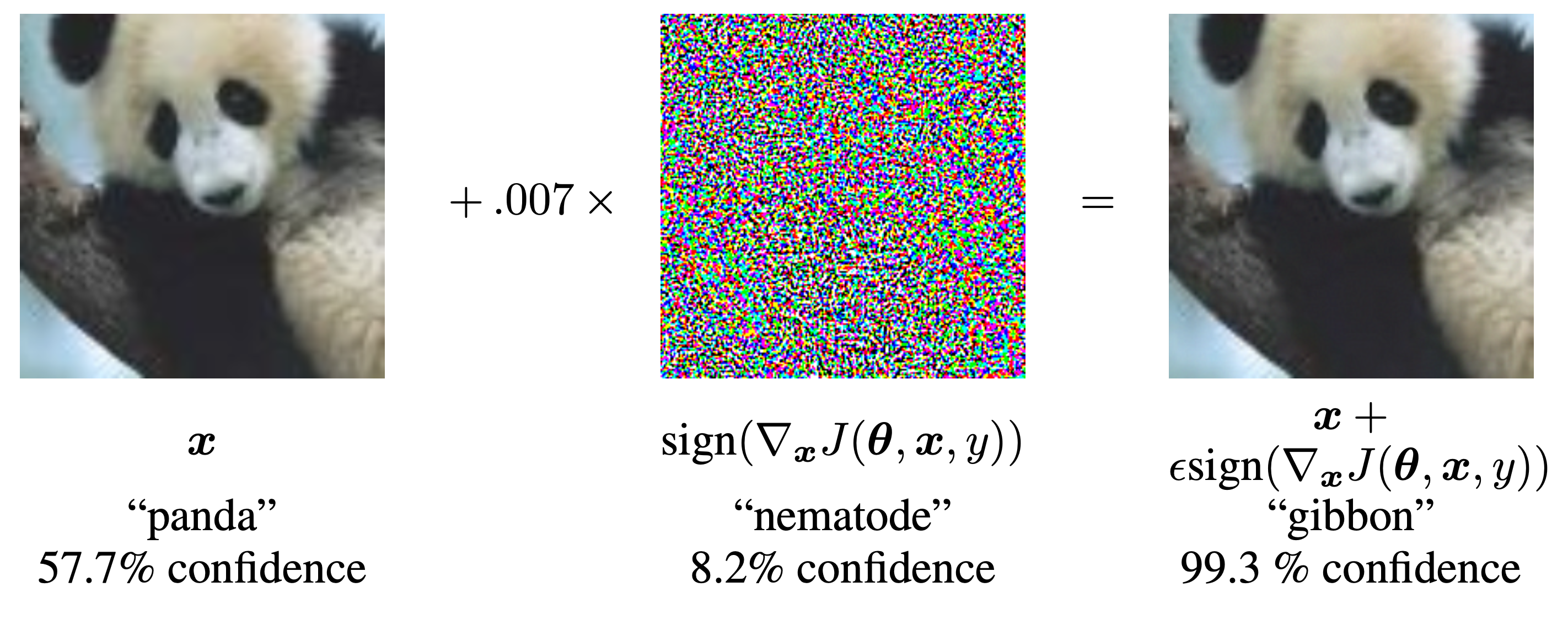
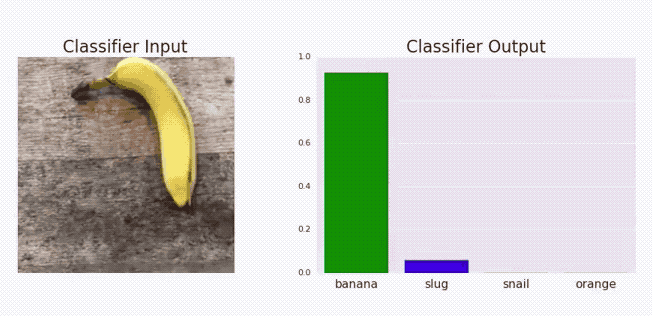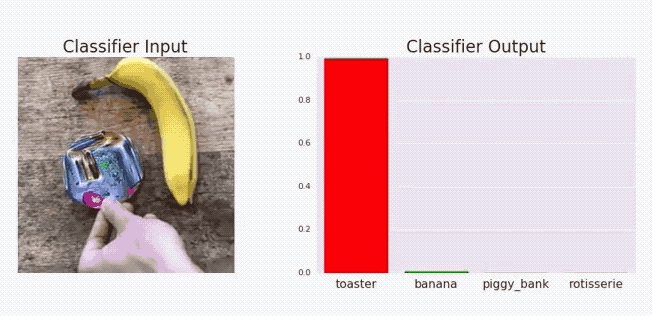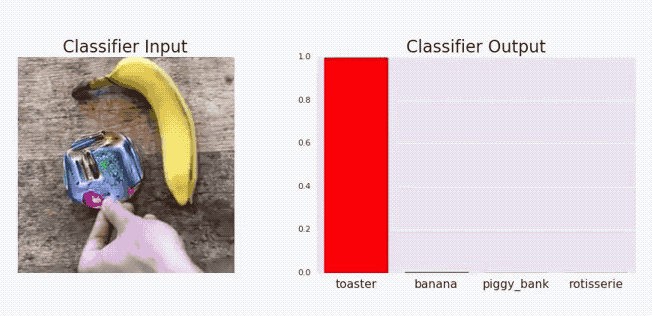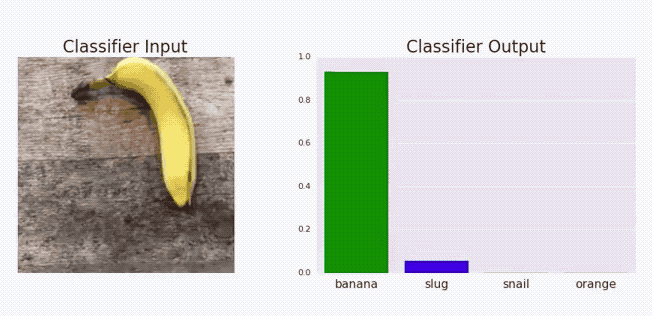
Goodfellow, Ian J, Jonathon Shlens, and Christian Szegedy. 2014. “Explaining and Harnessing Adversarial Examples.” arXiv Preprint arXiv:1412.6572.
Li, Cheng, Jindong Wang, Yixuan Zhang, Kaijie Zhu, Wenxin Hou, Jianxun Lian, Fang Luo, Qiang Yang, and Xing Xie. 2023. “Large Language Models Understand and Can Be Enhanced by Emotional Stimuli.” arXiv Preprint arXiv:2307.11760.
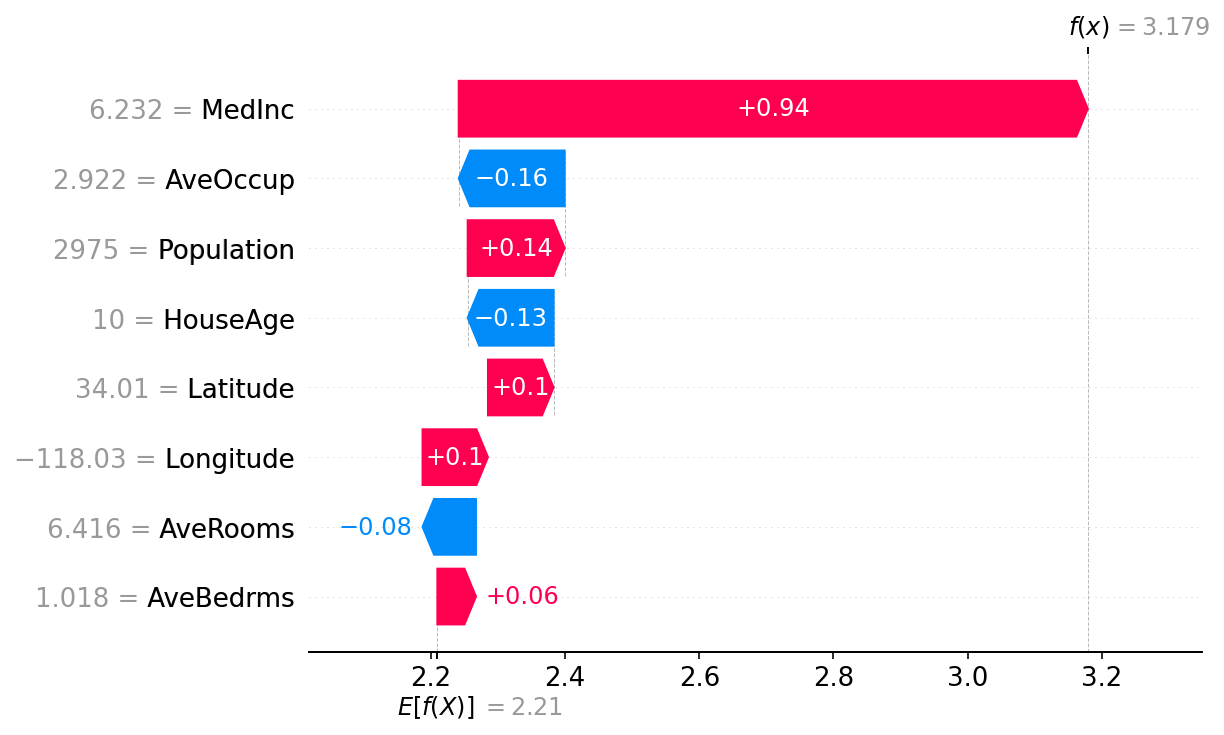
Lundberg, Scott M, and Su-In Lee. 2017. “A Unified Approach to Interpreting Model Predictions.” Advances in Neural Information Processing Systems 30.
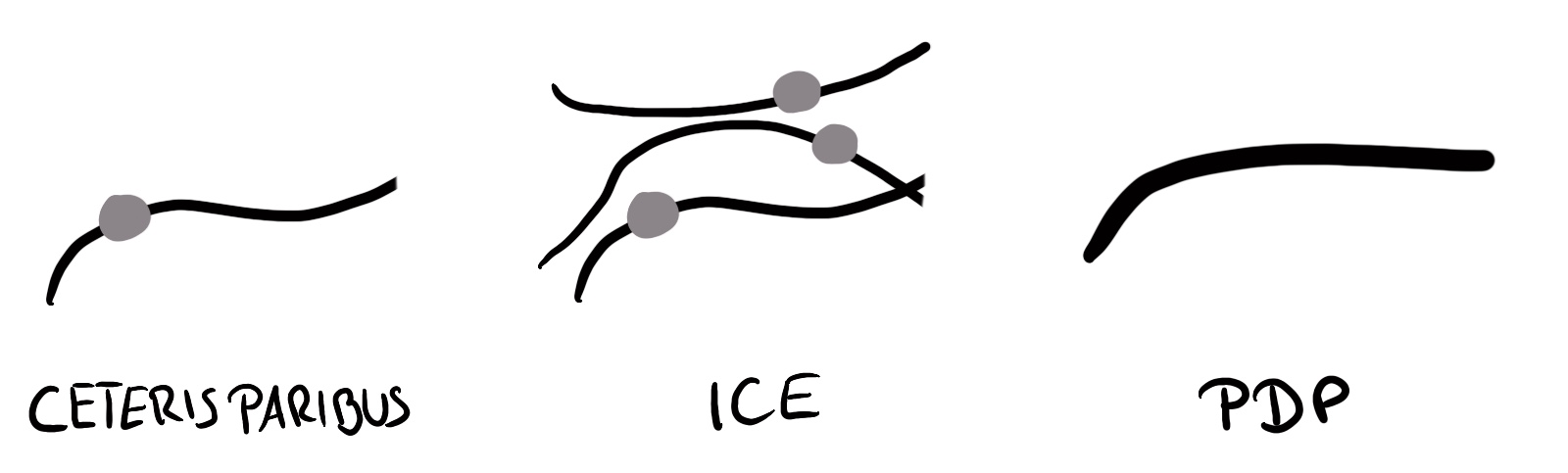
Molnar, Christoph. 2020. Interpretable Machine Learning.
Ribeiro, Marco Tulio, Sameer Singh, and Carlos Guestrin. 2016. “Why Should I Trust You?": Explaining the Predictions of Any Classifier.” In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 1135–44. Association for Computing Machinery.
Richman, Ronald, and Mario V. Wüthrich. 2023. “LocalGLMnet: Interpretable Deep Learning for Tabular Data.” Scandinavian Actuarial Journal 2023 (1): 71–95.
Rudin, Cynthia. 2019. “Stop Explaining Black Box Machine Learning Models for High Stakes Decisions and Use Interpretable Models Instead.” Nature Machine Intelligence 1 (5): 206–15.
Rudin, Cynthia, Chaofan Chen, Zhi Chen, Haiyang Huang, Lesia Semenova, and Chudi Zhong. 2022. “Interpretable Machine Learning: Fundamental Principles and 10 Grand Challenges.” Statistic Surveys 16: 1–85.